Research
A comparative evaluation of diffusion based networks for Multiple Sclerosis lesion segmentation
 A. Rondinella, F. Guarnera, A. Ortis, E. Crispino, G. Russo, F. Pappalardo, S. Battiato, “A comparative evaluation of diffusion based networks for Multiple Sclerosis lesion segmentation”, Submitted to BMC Bioinformatics (2024).
A. Rondinella, F. Guarnera, A. Ortis, E. Crispino, G. Russo, F. Pappalardo, S. Battiato, “A comparative evaluation of diffusion based networks for Multiple Sclerosis lesion segmentation”, Submitted to BMC Bioinformatics (2024).
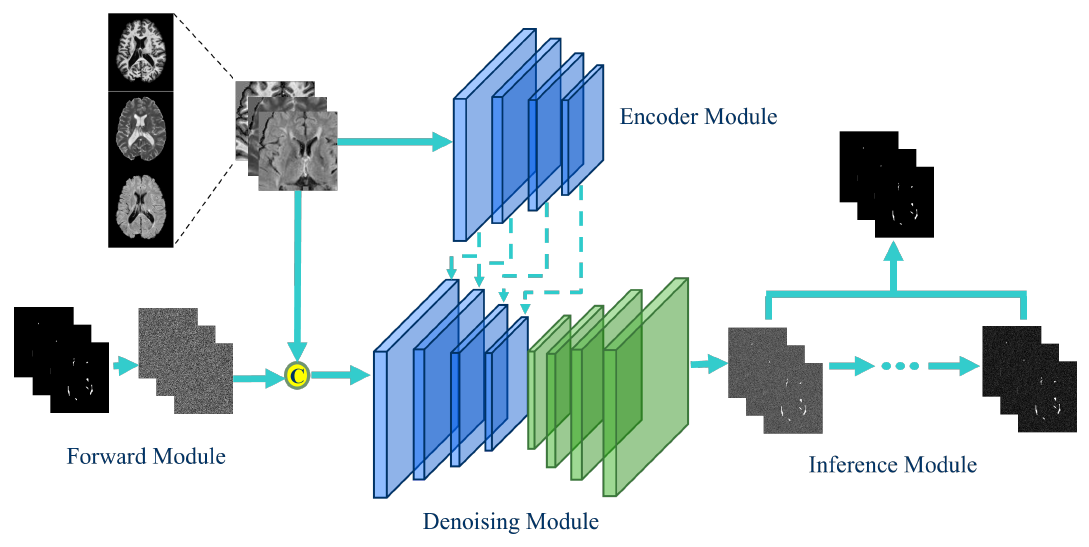
Semantic segmentation of Multiple Sclerosis (MS) lesions from longitudinal Magnetic Resonance Imaging (MRI) scans is crucial for the diagnosis and monitoring of disease progression. This study aims to evaluate the generalization performance of various deep learning segmentation models, which are commonly used in state-of-the-art medical image segmentation, when integrated into a diffusion model pipeline for segmenting MS lesions. Through an extensive set of experiments, we assess the performance of diffusion models with different architectural configurations to identify the optimal model for MS lesion segmentation. Additionally, we explored the robustness of diffusion model predictions by implementing various inference strategies to combine the diffusion model outputs obtained at each time step. Our results demonstrate the effectiveness of certain backbone architectures in enhancing diffusion model performance in MS lesion segmentation. Moreover, we demonstrate that accurate selection of inference strategies can further enhance the accuracy and robustness of diffusion model predictions. This study contributes to advancing the understanding of diffusion models’ applicability in clinical settings and provides insights for improving MS lesion segmentation in MRI. Our source code is freely available at https://github.com/alessiarondinella/MSSegDiff .
Advantages of brain parcellation in Multiple Sclerosis Lesion Segmentation
 D. Pishvai, A. Rondinella, F. Guarnera, S. Battiato, “Advantages of brain parcellation in Multiple Sclerosis Lesion Segmentation”, 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM).
D. Pishvai, A. Rondinella, F. Guarnera, S. Battiato, “Advantages of brain parcellation in Multiple Sclerosis Lesion Segmentation”, 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM).
Segmentation of multiple sclerosis lesions plays an important role in understanding disease status. In this work, we focus on the effectiveness of brain parcellation in enhancing the performance of segmentation for multiple sclerosis lesions in Magnetic Resonance Imaging. Brain parcellation does not improve the segmentation performance, but make the results more robust in terms of overall variability (e.g. standard deviation), by dividing the brain into physically significant sub-regions that the model can concentrate on. Our approach combines parcellation with the existing diffusion-based model to increase sensitivity, particularly in regions with small anomalies. We conducted a thorough evaluation of a reference dataset on the field using all available modalities. Our results show how the parcellation of the brain when integrated into a diffusion-based pipeline, makes the segmentation of MS more stable, lowering deviations from the average, and improving some of the results w.r.t. state-of-theart. This method achieves good segmentation capabilities even with small datasets, providing promising indications for further research.
ICPR 2024 Competition on Multiple Sclerosis Lesion Segmentation - Methods and Results
 A. Rondinella, F. Guarnera, E. Crispino, G. Russo, C. Di Lorenzo, D. Maimone, F. Pappalardo, S. Battiato, “ICPR 2024 Competition on Multiple Sclerosis Lesion Segmentation – Methods and Results”. 2024 International Conference on Pattern Recognition (ICPR 2024).
Competition Link
A. Rondinella, F. Guarnera, E. Crispino, G. Russo, C. Di Lorenzo, D. Maimone, F. Pappalardo, S. Battiato, “ICPR 2024 Competition on Multiple Sclerosis Lesion Segmentation – Methods and Results”. 2024 International Conference on Pattern Recognition (ICPR 2024).
Competition Link
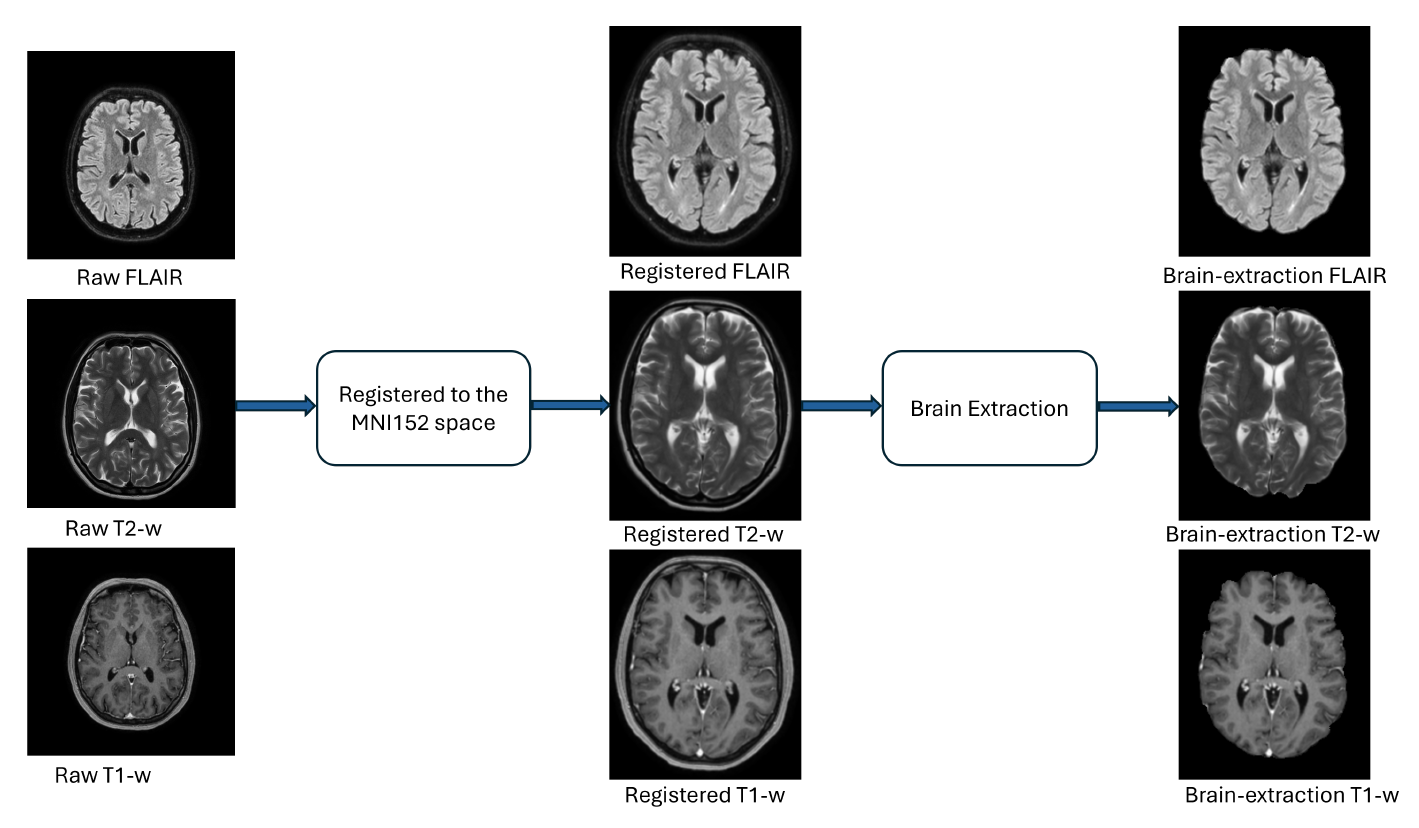
This report summarizes the outcomes of the ICPR 2024 Competition on Multiple Sclerosis Lesion Segmentation (MSLesSeg). The competition aimed to develop methods capable of automatically segmenting multiple sclerosis lesions in MRI scans. Participants were provided with a novel annotated dataset comprising a heterogeneous cohort of MS patients, featuring both baseline and follow-up MRI scans acquired at different hospitals. MSLesSeg focuses on developing algorithms that can independently segment multiple sclerosis lesions of an unexamined cohort of patients. This segmentation approach aims to overcome current benchmarks by eliminating user interaction and ensuring robust lesion detection at different timepoints, encouraging innovation and promoting methodological advances.
SynthBA: Reliable Brain Age Estimation Across Multiple MRI Sequences and Resolutions
 L. Puglisi, A. Rondinella, L. De Meo, F. Guarnera, S. Battiato, D. Ravì, “SynthBA: Reliable Brain Age Estimation Across Multiple MRI Sequences and Resolutions”. MetroXRAINE 2024.
L. Puglisi, A. Rondinella, L. De Meo, F. Guarnera, S. Battiato, D. Ravì, “SynthBA: Reliable Brain Age Estimation Across Multiple MRI Sequences and Resolutions”. MetroXRAINE 2024.
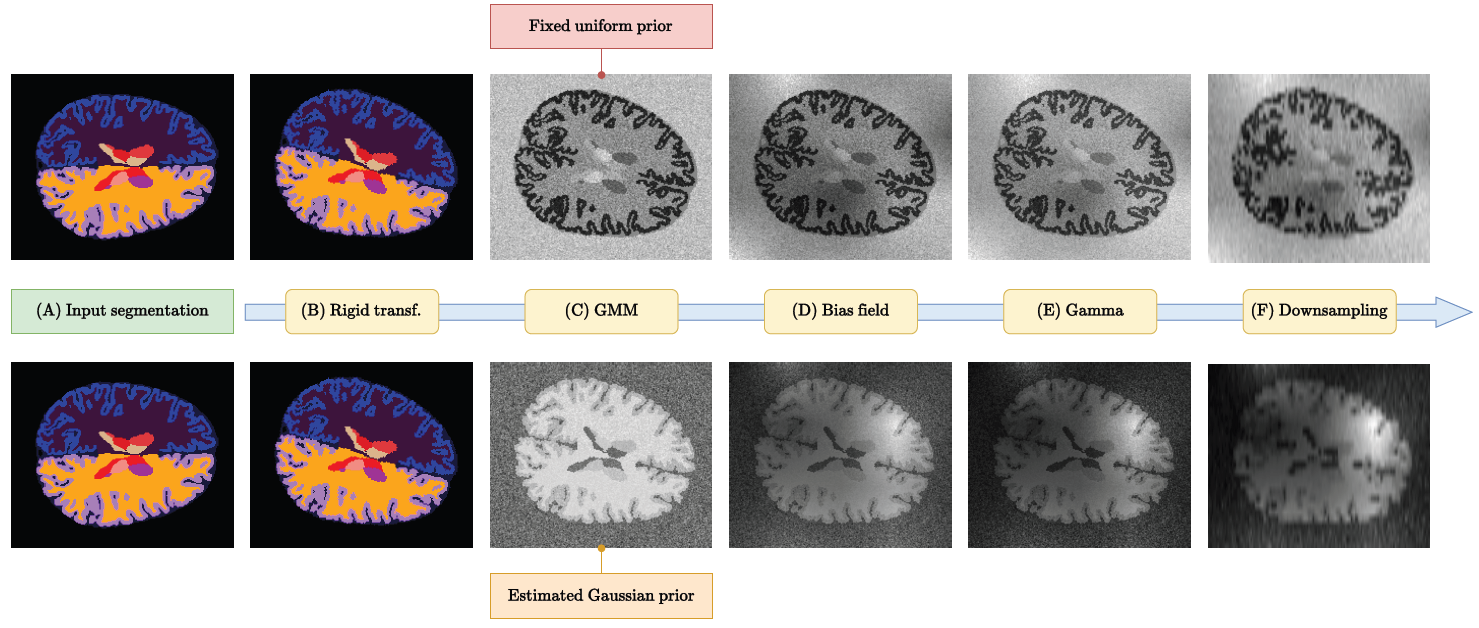
Brain age is a critical measure that reflects the biological ageing process of the brain. The gap between brain age and chronological age, referred to as brain PAD (Predicted Age Difference), has been utilized to investigate neurodegenerative conditions. Brain age can be predicted using MRIs and machine learning techniques. However, existing methods are often sensitive to acquisition-related variabilities, such as differences in acquisition protocols, scanners, MRI sequences, and resolutions, significantly limiting their application in highly heterogeneous clinical settings. In this study, we introduce Synthetic Brain Age (SynthBA), a robust deep-learning model designed for predicting brain age. SynthBA utilizes an advanced domain randomization technique, ensuring effective operation across a wide array of acquisition-related variabilities. To assess the effectiveness and robustness of SynthBA, we evaluate its predictive capabilities on internal and external datasets, encompassing various MRI sequences and resolutions, and compare it with state-of-the-art techniques. Additionally, we calculate the brain PAD in a large cohort of subjects with Alzheimer’s Disease (AD), demonstrating a significant correlation with AD-related measures of cognitive dysfunction. SynthBA holds the potential to facilitate the broader adoption of brain age prediction in clinical settings, where re-training or fine-tuning is often unfeasible. The SynthBA source code and pre-trained models are publicly available at https://github.com/LemuelPuglisi/SynthBA.
Efficient Atrophy Mapping: A Single-Step U-net Approach for Rapid Brain Change Estimation
 R. Raciti, A. Rondinella, L. Puglisi, F. Guarnera, D. Ravì, S. Battiato, “Efficient Atrophy Mapping: A Single-Step U-net Approach for Rapid Brain Change Estimation”. MetroXRAINE 2024
R. Raciti, A. Rondinella, L. Puglisi, F. Guarnera, D. Ravì, S. Battiato, “Efficient Atrophy Mapping: A Single-Step U-net Approach for Rapid Brain Change Estimation”. MetroXRAINE 2024
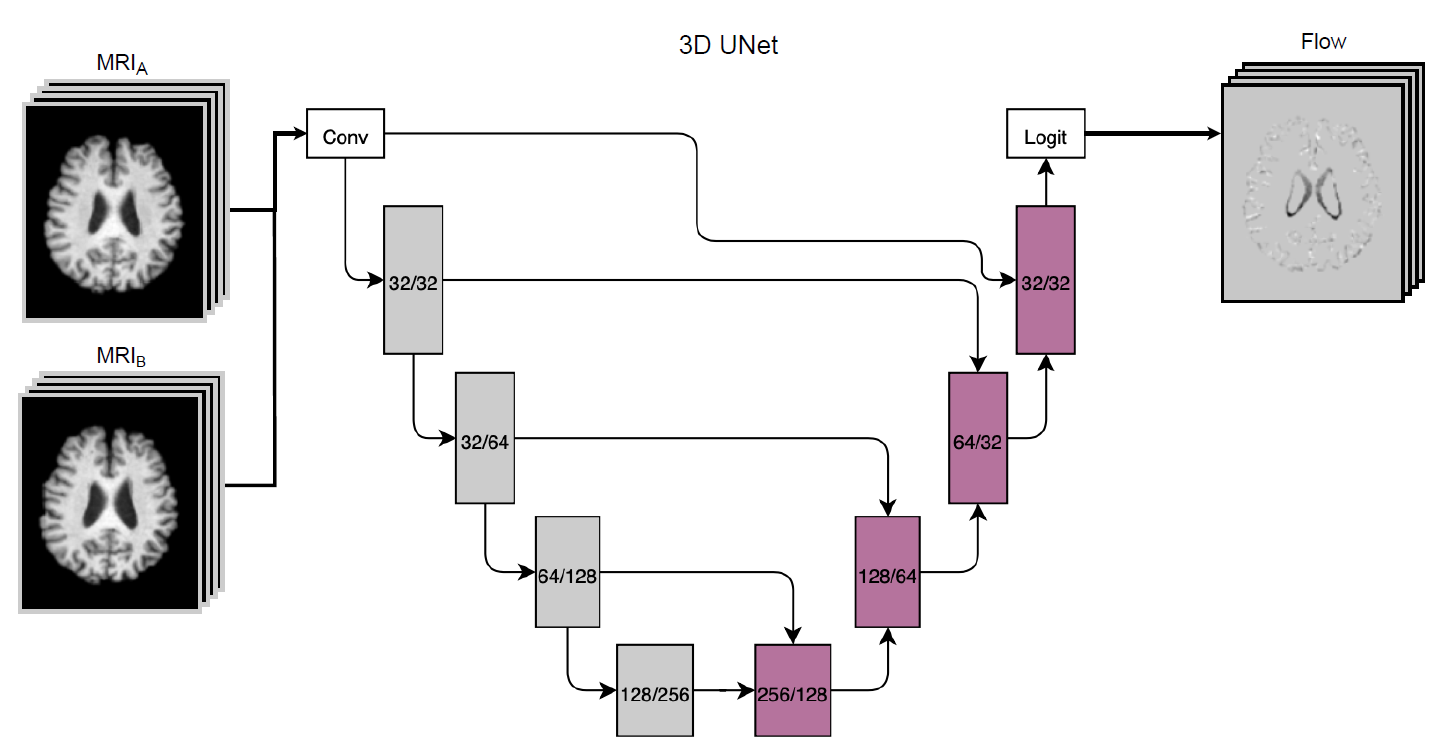
The estimation of brain atrophy is crucial for evaluating brain diseases and analyzing neurodegeneration. Existing methods for computing atrophy maps often suffer from lengthy processing times due to the computational cost of multi-step processing. In this work, we propose a novel technique for atrophy map calculation using a single U-net-based architecture.This approach consolidates multiple traditional medical imaging processing steps into a single process, aiming to accelerate the computational time required. Specifically, our method estimates structural changes by generating a flow map from two longitudinal Magnetic Resonance Imaging (MRI) scans of the same subject. We trained and evaluated our system on a dataset comprising 2000 T1-weighted MRI scans sourced from two different public datasets on Alzheimer’s Disease. Experimental results demonstrate a considerable reduction in execution time while maintaining atrophy mapping performance comparable to state-of-the-art solutions. We believe that our pipeline could significantly benefit clinical applications for measuring brain atrophy, especially in scenarios requiring the evaluation of large cohorts, such as clinical trials. Our code is freely available at https://github.com/Raciti/Efficient-Atrophy-Mapping-A-Single-Step-U-net-Approach-for-Rapid-Brain-Change-Estimation.
TADM: Temporally-Aware Diffusion Model for Neurodegenerative Progression on Brain MRI
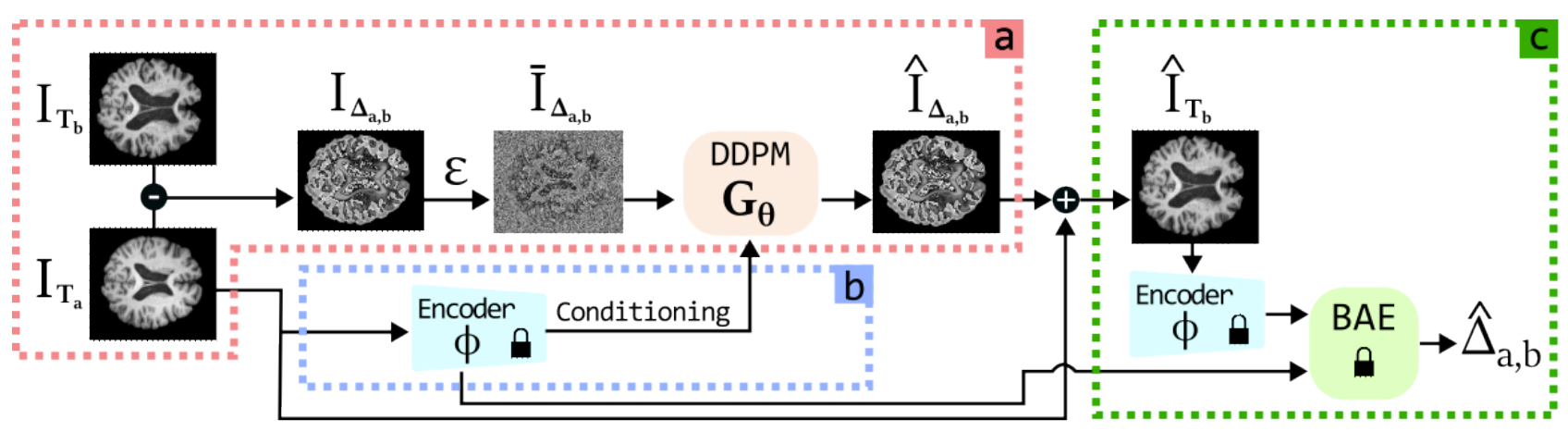 M. Litrico, F. Guarnera, M.V. Giuffrida, D. Ravì, S. Battiato, "TADM: Temporally-Aware Diffusion Model for Neurodegenerative Progression on Brain MRI." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2024.
M. Litrico, F. Guarnera, M.V. Giuffrida, D. Ravì, S. Battiato, "TADM: Temporally-Aware Diffusion Model for Neurodegenerative Progression on Brain MRI." International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer Nature Switzerland, 2024.
Generating realistic images to accurately predict changes in the structure of brain MRI can be a crucial tool for clinicians. Such applications can help assess patients’ outcomes and analyze how diseases progress at the individual level. However, existing methods developed for this task present some limitations. Some approaches attempt to model the distribution of MRI scans directly by conditioning the model on patients’ ages, but they fail to explicitly capture the relationship between structural changes in the brain and time intervals, especially on age-unbalanced datasets. Other approaches simply rely on interpolation between scans, which limits their clinical application as they do not predict future MRIs. To address these challenges, we propose a Temporally-Aware Diffusion Model (TADM), which introduces a novel approach to accurately infer progression in brain MRIs. TADM learns the distribution of structural changes in terms of intensity differences between scans and combines the prediction of these changes with the initial baseline scans to generate future MRIs. Furthermore, during training, we propose to leverage a pre-trained Brain-Age Estimator (BAE) to refine the model’s training process, enhancing its ability to produce accurate MRIs that match the expected age gap between baseline and generated scans. Our assessment, conducted on 634 subjects from the OASIS-3 dataset, uses similarity metrics and region sizes computed by comparing predicted and real follow-up scans on 3 relevant brain regions. TADM achieves large improvements over existing approaches, with an average decrease of 24% in region size error and an improvement of 4% in similarity metrics. These evaluations demonstrate the improvement of our model in mimicking temporal brain neurodegenerative progression compared to existing methods. We believe that our approach will significantly benefit clinical applications, such as predicting patient outcomes or improving treatments for patients. Our code is publicly available at https://github.com/MattiaLitrico/TADM-Temporally-Aware-Diffusion-Model-for-Neurodegenerative-Progression-on-Brain-MRI.
Enhancing Multiple Sclerosis Lesion Segmentation in Multimodal MRI Scans with Diffusion Models
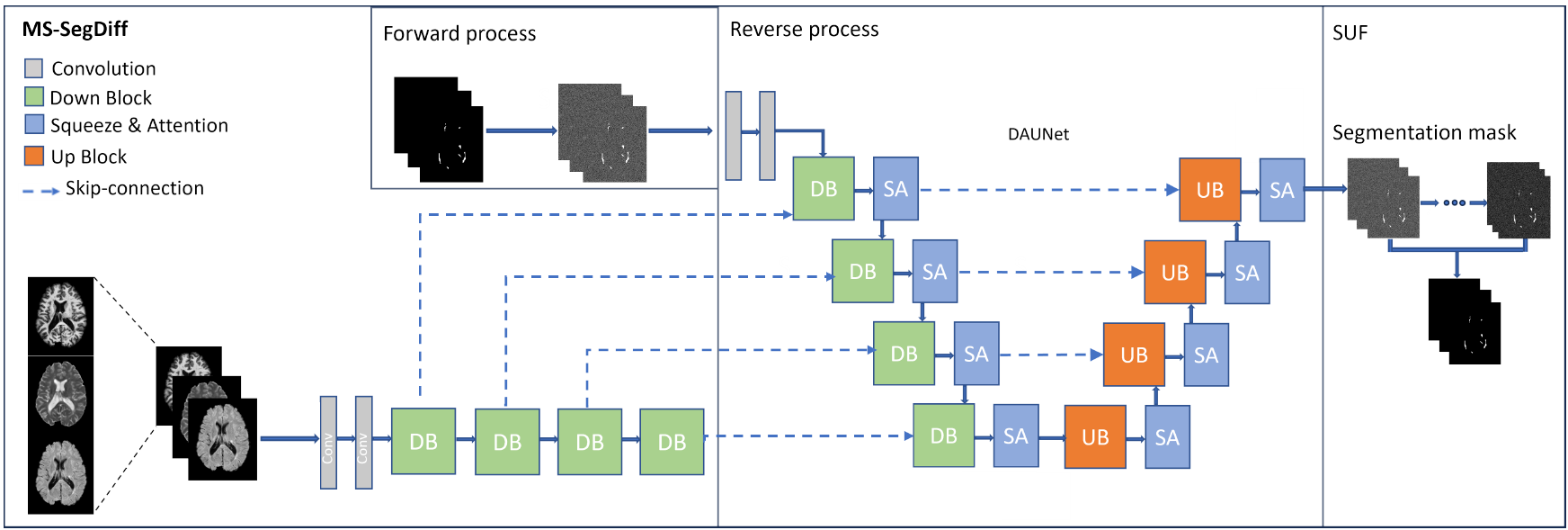 A. Rondinella, F. Guarnera, O. Giudice, A. Ortis, G. Russo, E. Crispino, F. Pappalardo, S. Battiato. "Enhancing Multiple Sclerosis Lesion Segmentation in Multimodal MRI Scans with Diffusion Models." 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2023.
A. Rondinella, F. Guarnera, O. Giudice, A. Ortis, G. Russo, E. Crispino, F. Pappalardo, S. Battiato. "Enhancing Multiple Sclerosis Lesion Segmentation in Multimodal MRI Scans with Diffusion Models." 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2023.
Accurate segmentation of Multiple Sclerosis (MS) lesions from Magnetic Resonance Imaging (MRI) scans is crucial for clinical diagnosis and effective treatment planning. In this work, we investigate the effectiveness of Diffusion Models (DM) in achieving pixel-wise segmentation of MS lesions. DM significantly improves segmentation sensitivity, especially in regions with subtle abnormalities. We conducted extensive experiments using the magnetic resonance volumes from a public dataset, encompassing various imaging modalities. Our analysis demonstrated how DM can achieve performance levels that are on par with state-of-the-art techniques, as evidenced by a mean Dice coefficient comparable to the best existing methods. Furthermore, some variants of standard DM exhibits robustness across various imaging modalities, showcasing its versatility in clinical settings.
Boosting multiple sclerosis lesion segmentation through attention mechanism
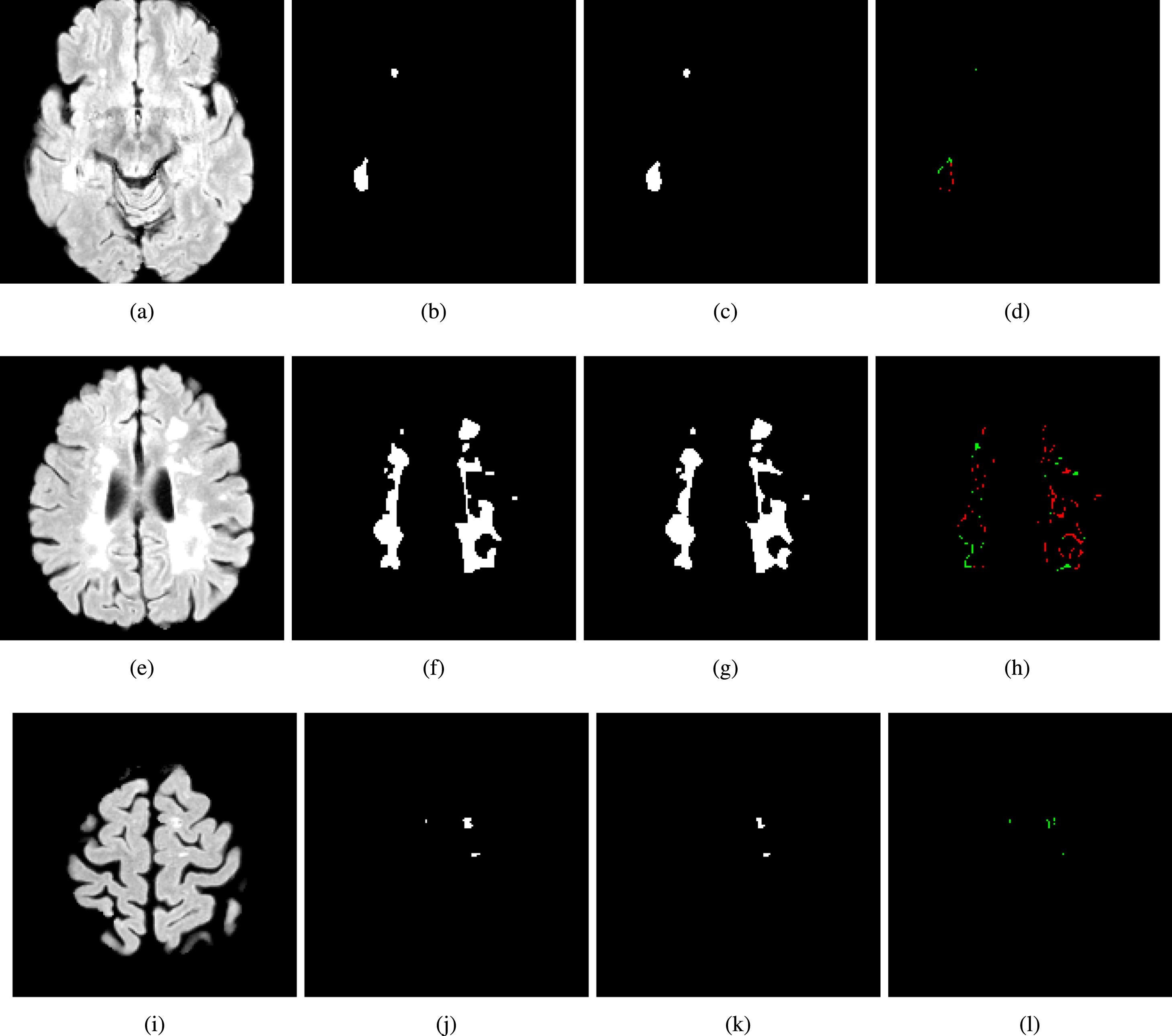 A. Rondinella, E. Crispino, F. Guarnera, O. Giudice, A. Ortis, G. Russo, C. Di Lorenzo, D. Maimone, F. Pappalardo, S. Battiato. Boosting multiple sclerosis lesion segmentation through attention mechanism. Computers in Biology and Medicine, 2023, 107021.
A. Rondinella, E. Crispino, F. Guarnera, O. Giudice, A. Ortis, G. Russo, C. Di Lorenzo, D. Maimone, F. Pappalardo, S. Battiato. Boosting multiple sclerosis lesion segmentation through attention mechanism. Computers in Biology and Medicine, 2023, 107021.
Magnetic resonance imaging is a fundamental tool to reach a diagnosis of multiple sclerosis and monitoring its progression. Although several attempts have been made to segment multiple sclerosis lesions using artificial intelligence, fully automated analysis is not yet available. State-of-the-art methods rely on slight variations in segmentation architectures (e.g. U-Net, etc.). However, recent research has demonstrated how exploiting temporal-aware features and attention mechanisms can provide a significant boost to traditional architectures. This paper proposes a framework that exploits an augmented U-Net architecture with a convolutional long short-term memory layer and attention mechanism which is able to segment and quantify multiple sclerosis lesions detected in magnetic resonance images. Quantitative and qualitative evaluation on challenging examples demonstrated how the method outperforms previous state-of-the-art approaches, reporting an overall Dice score of 89% and also demonstrating robustness and generalization ability on never seen new test samples of a new dedicated under construction dataset.
Attention-Based Convolutional Neural Network for CT Scan COVID-19 Detection
 A. Rondinella, F. Guarnera, O. Giudice, A. Ortis, F. Rundo, S. Battiato. Attention-Based Convolutional Neural Network for CT Scan COVID-19 Detection. Proceedings of the IEEE International Conference on Acoustics Speech and Signal Processing Satellite Workshops, IEEE (2023).
A. Rondinella, F. Guarnera, O. Giudice, A. Ortis, F. Rundo, S. Battiato. Attention-Based Convolutional Neural Network for CT Scan COVID-19 Detection. Proceedings of the IEEE International Conference on Acoustics Speech and Signal Processing Satellite Workshops, IEEE (2023).
The accurate detection of Covid-19 from chest Computed Tomography (CT) images can assist in early diagnosis and management of the disease. This paper presents a solution for Covid-19 detection, presented in the challenge of 3rd Covid-19 competition, inside the “AI-enabled Medical Image Analysis Workshop” organized by IEEE International Conference on Acoustic, Speech and Signal Processing (ICASSP) 2023. In this work, the application of deep learning models for chest CT image analysis was investigated, focusing on the use of a ResNet as a backbone network augmented with attention mechanisms. The ResNet provides an effective feature extractor for the classification task, while the attention mechanisms improve the model’s ability to focus on important regions of interest within the images. We conducted extensive experiments on a provided dataset and achieved a macro F1 score of 0.78 on the test set, demonstrating the potential to assist the diagnosis of Covid-19. Our proposed approach leverages the power of deep learning with attention mechanisms to address the challenges of Covid-19 detection in the early detection and management of the disease. In both test and validation set, the proposed method outperformed the baseline of the challenge, ranking fifth in the competition.
Early Detection of Hip Periprosthetic Joint Infections Through CNN on Computed Tomography Images
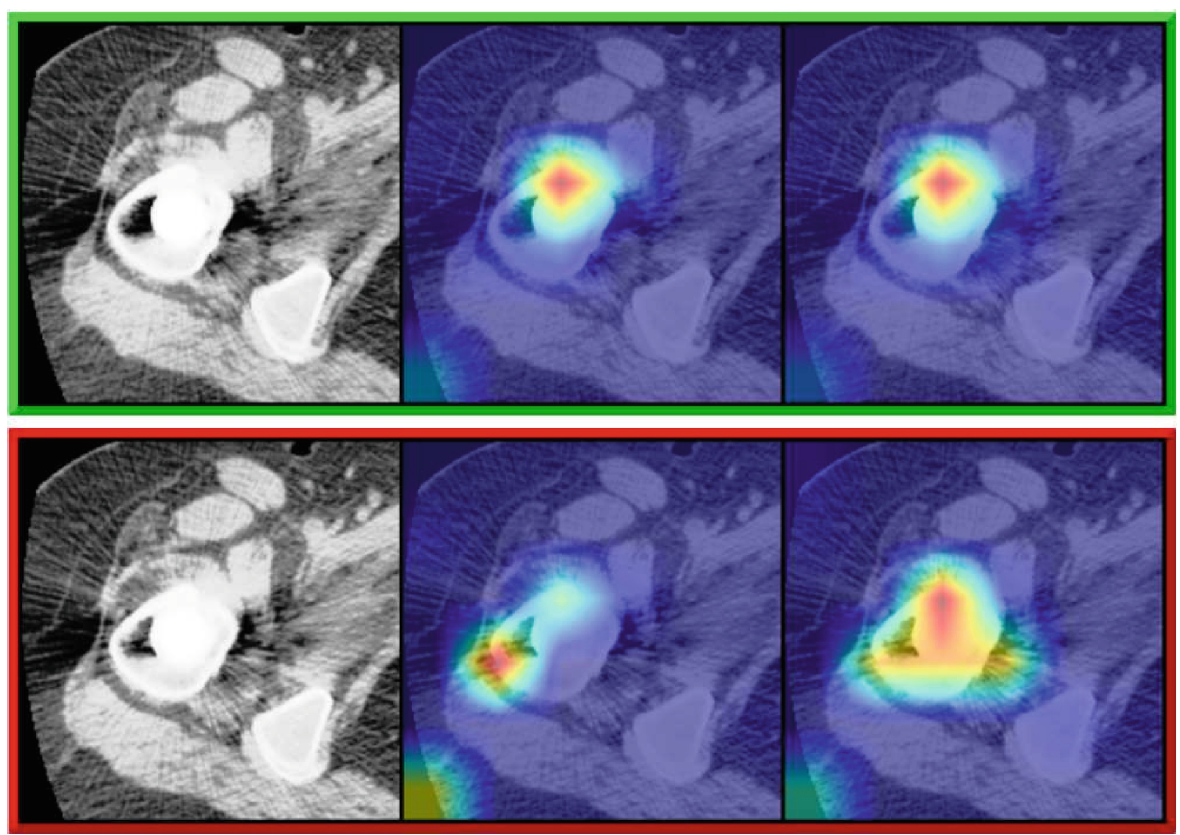 F. Guarnera, A. Rondinella, O. Giudice, A. Ortis, S. Battiato, F. Rundo, G. Fallica, F. Traina, S. Conoci. "Early detection of hip periprosthetic joint infections through cnn on computed tomography images." International Conference on Image Analysis and Processing. Cham: Springer Nature Switzerland, 2023.
F. Guarnera, A. Rondinella, O. Giudice, A. Ortis, S. Battiato, F. Rundo, G. Fallica, F. Traina, S. Conoci. "Early detection of hip periprosthetic joint infections through cnn on computed tomography images." International Conference on Image Analysis and Processing. Cham: Springer Nature Switzerland, 2023.
Early detection of an infection prior to prosthesis removal (e.g., hips, knees or other areas) would provide significant benefits to patients. Currently, the detection task is carried out only retrospectively with a limited number of methods relying on biometric or other medical data. The automatic detection of a periprosthetic joint infection from tomography imaging is a task never addressed before. This study introduces a novel method for early detection of the hip prosthesis infections analyzing Computed Tomography images. The proposed solution is based on a novel ResNeSt Convolutional Neural Network architecture trained on samples from more than 100 patients. The solution showed exceptional performance in detecting infections with an experimental high level of accuracy and F-score.
Mixup Data Augmentation for COVID-19 Infection Percentage Estimation
Napoli Spatafora, M. A., Ortis, A., & Battiato, S. (2022, August). Mixup Data Augmentation for COVID-19 Infection Percentage Estimation. In Image Analysis and Processing. ICIAP 2022 Workshops: ICIAP International Workshops, Lecce, Italy, May 23–27, 2022, Revised Selected Papers, Part II (pp. 508-519). Cham: Springer International Publishing.
The outbreak of the COVID-19 pandemic considerably increased the workload in hospitals. In this context, the availability of proper diagnostic tools is very important in the fight against this virus. Scientific research is constantly making its contribution in this direction. Actually, there are many scientific initiatives including challenges that require to develop deep algorithms that analyse X-ray or Computer Tomography (CT) images of lungs. One of these concerns a challenge whose topic is the prediction of the percentage of COVID-19 infection in chest CT images. In this paper, we present our contribution to the COVID-19 Infection Percentage Estimation Competition organised in conjunction with the ICIAP 2021 Conference. The proposed method employs algorithms for classification problems such as Inception-v3 and the technique of data augmentation mixup on COVID-19 images. Moreover, the mixup methodology is applied for the first time in radiological images of lungs affected by COVID-19 infection, with the aim to infer the infection degree with slice-level precision. Our approach achieved promising results despite the specific constrains defined by the rules of the challenge, in which our solution entered in the final ranking.
An Explainable Medical Imaging Framework for Modality Classifications Trained Using Small Datasets
Trenta, F., Battiato, S., & Ravì, D. (2022, May). An Explainable Medical Imaging Framework for Modality Classifications Trained Using Small Datasets. In Image Analysis and Processing–ICIAP 2022: 21st International Conference, Lecce, Italy, May 23–27, 2022, Proceedings, Part I (pp. 358-367). Cham: Springer International Publishing.
With the huge expansion of artificial intelligence in medical imaging, many clinical warehouses, medical centres and research communities, have organized patients’ data in well-structured datasets. These datasets are one of the key elements to train AI-enabled solutions. Additionally, the value of such datasets depends on the quality of the underlying data. To maintain the desired high-quality standard, these datasets are actively cleaned and continuously expanded. This labelling process is time-consuming and requires clinical expertise even when a simple classification task must be performed. Therefore, in this work, we propose to tackle this problem by developing a new pipeline for the modality classification of medical images. Our pipeline has the purpose to provide an initial step in organizing a large collection of data and grouping them by modality, thus reducing the involvement of costly human raters. In our experiments, we consider 4 popular deep neural networks as the core engine of the proposed system. The results show that when limited datasets are available simpler pre-trained networks achieved better results than more complex and sophisticated architectures. We demonstrate this by comparing the considered networks on the ADNI dataset and by exploiting explainable AI techniques that help us to understand our hypothesis. Still today, many medical imaging studies make use of limited datasets, therefore we believe that our contribution is particularly relevant to drive future developments of new medical imaging technologies when limited data are available.
Advanced Deep Network with Attention and Genetic-Driven Reinforcement Learning Layer for an Efficient Cancer Treatment Outcome Prediction
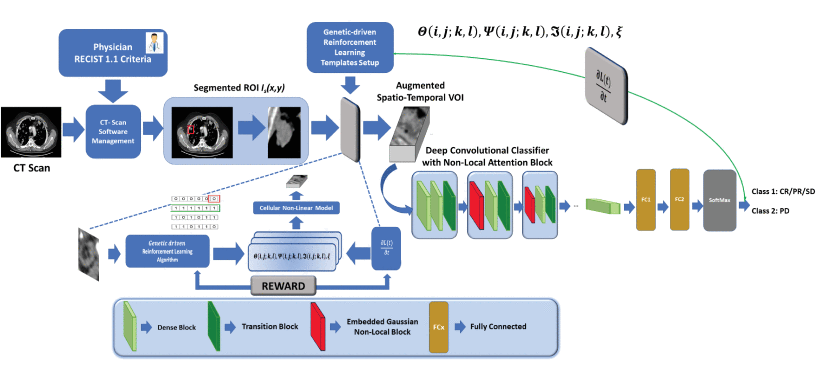 F. Rundo, G. L. Banna, F. Trenta and S. Battiato, "Advanced Deep Network with Attention and Genetic-Driven Reinforcement Learning Layer for an Efficient Cancer Treatment Outcome Prediction," 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 2021, pp. 294-298, doi: 10.1109/ICIP42928.2021.9506805.
F. Rundo, G. L. Banna, F. Trenta and S. Battiato, "Advanced Deep Network with Attention and Genetic-Driven Reinforcement Learning Layer for an Efficient Cancer Treatment Outcome Prediction," 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 2021, pp. 294-298, doi: 10.1109/ICIP42928.2021.9506805.
In the last few years, medical researchers have investigated promising approaches for cancer treatment, leading to a major interest in the immunotherapeutic approach. The target of immunotherapy is to boost a subject’s immune system in order to fight cancer. However, scientific studies confirmed that not all patients have a positive response to immunotherapy treatment. Medical research has long been engaged in the search for predictive immunotherapeutic-response biomarkers. Based on these considerations, we developed a non-invasive advanced pipeline with a downstream 3D deep classifier with attention and reinforcement learning for early prediction of patients responsive to immunotherapeutic treatment from related chest-abdomen CT-scan imaging. We have tested the proposed pipeline within a clinical trial that recruited patients with metastatic bladder cancer. Our experiment results achieved accuracy close to 93%.
Advanced Densely Connected System with Embedded Spatio-Temporal Features Augmentation for Immunotherapy Assessment
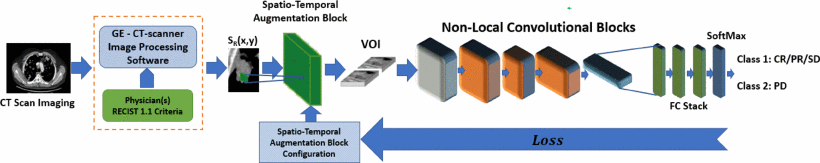 F. Rundo, G. L. Banna, F. Trenta and S. Battiato, "Advanced Densely Connected System with Embedded Spatio-Temporal Features Augmentation for Immunotherapy Assessment," 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 2021, pp. 1-7, doi: 10.1109/IJCNN52387.2021.9534078.
F. Rundo, G. L. Banna, F. Trenta and S. Battiato, "Advanced Densely Connected System with Embedded Spatio-Temporal Features Augmentation for Immunotherapy Assessment," 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 2021, pp. 1-7, doi: 10.1109/IJCNN52387.2021.9534078.
In medical field, the term “immunotherapy” refers to a form of cancer treatment that uses the ability of body's immune system to prevent and destroy cancer cells. In the last few years, immunotherapy has demonstrated to be a very effective treatment in fighting cancer diseases. However, immunotherapy does not work for every patients and moreover, certain types of immunotherapy drugs could have side effects. With this regard, scientific researchers are investigating for effective ways to select the patients who are more likely to respond to the treatment. Hence, pre-clinical data confirmed that, sometimes, the composition of immune system cells infiltrating the tumor micro-environment may interfere with the efficacy of immunotherapy treatments. In this work, we developed a 3D Deep Network with a downstream classifier for selecting and properly augmenting features from chest-abdomen CT images toward improving cancer outcome prediction. In our work, we proposed an effective solution to a specific type of aggressive bladder cancer, called Metastatic Urothelial Carcinoma (mUC). Our experiment results achieved high accuracy confirming the effectiveness of the proposed pipeline.
Interpretable Deep Model For Predicting Gene-Addicted Non-Small-Cell Lung Cancer In Ct Scans
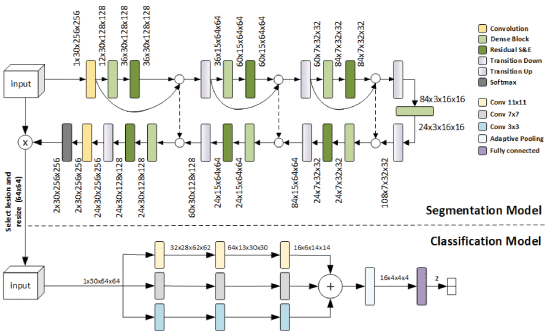 Pino, C., Palazzo, S., Trenta, F., Cordero, F., Bagci, U., Rundo, F., Battiato, S., Giordano, D., Aldinucci, M., & Spampinato, C. (2021, April). Interpretable deep model for predicting gene-addicted non-small-cell lung cancer in ct scans. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) (pp. 891-894). IEEE.
Pino, C., Palazzo, S., Trenta, F., Cordero, F., Bagci, U., Rundo, F., Battiato, S., Giordano, D., Aldinucci, M., & Spampinato, C. (2021, April). Interpretable deep model for predicting gene-addicted non-small-cell lung cancer in ct scans. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) (pp. 891-894). IEEE.
Genetic profiling and characterization of lung cancers have recently emerged as a new technique for targeted therapeutic treatment based on immunotherapy or molecular drugs. However, the most effective way to discover specific gene mutations through tissue biopsy has several limitations, from invasiveness to being a risky procedure. Recently, quantitative assessment of visual features from CT data has been demonstrated to be a valid alternative to biopsy for the diagnosis of gene-addicted tumors. In this paper, we present a deep model for automated lesion segmentation and classification as gene-addicted or not. The segmentation approach extends the 2D Tiramisu architecture for 3D segmentation through dense blocks and squeeze-and-excitation layers, while a multi-scale 3D CNN is used for lesion classification. We also train our model with adversarial samples, and show that this approach acts as a gradient regularizer and enhances model interpretability. We also built a dataset, the first of its nature, consisting of 73 CT scans annotated with the presence of a specific genomics profile. We test our approach on this dataset achieving a segmentation accuracy of 93.11% (Dice score) and a classification accuracy in identifying oncogene-addicted lung tumors of 82.00%.
Advanced 3D Deep Non-Local Embedded System for Self-Augmented X-Ray-Based COVID-19 Assessment
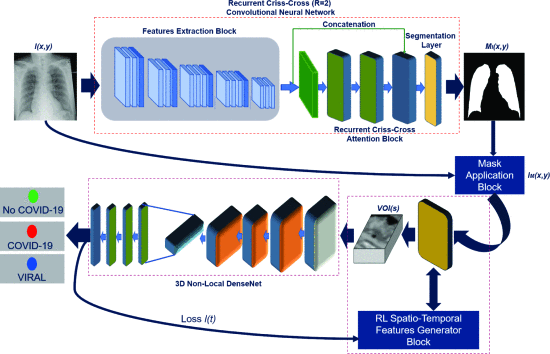 Rundo, F., Genovese, A., Leotta, R., Scotti, F., Piuri, V., & Battiato, S. (2021). Advanced 3d deep non-local embedded system for self-augmented x-ray-based covid-19 assessment. In Proceedings of the IEEE/CVF International
Conference on Computer Vision (pp. 423-432).
Rundo, F., Genovese, A., Leotta, R., Scotti, F., Piuri, V., & Battiato, S. (2021). Advanced 3d deep non-local embedded system for self-augmented x-ray-based covid-19 assessment. In Proceedings of the IEEE/CVF International
Conference on Computer Vision (pp. 423-432).
COVID-19 diagnosis using chest x-ray (CXR) imaging has a greater sensitivity and faster acquisition procedures than the Real-Time Polimerase Chain Reaction (RT-PCR) test, also requiring radiology machinery that is cheap and widely available. To process the CXR images, methods based on Deep Learning (DL) are being increasingly used, often in combination with data augmentation techniques. However, no method in the literature performs data augmentation in which the augmented training samples are processed collectively as a multi-channel image. Furthermore, no approach has yet considered a combination of attention-based networks with Convolutional Neural Networks (CNN) for COVID-19 detection. In this paper, we propose the first method for COVID-19 detection from CXR images that uses an innovative self-augmentation scheme based on reinforcement learning, which combines all the augmented images in a 3D deep volume and processes them together using a novel non-local deep CNN, which integrates convolutional and attention layers based on non-local blocks. Results on publicly-available databases exhibit a greater accuracy than the state of the art, also showing that the regions of CXR images influencing the decision are consistent with radiologists' observations.
Advanced Non-linear Generative Model with a Deep Classifier for Immunotherapy Outcome Prediction: A Bladder Cancer Case Study
Rundo, F., Banna, G. L., Trenta, F., Spampinato, C., Bidaut, L., Ye, X., Kollias, S., & Battiato, S. (2021). Advanced non-linear generative model with a deep classifier for immunotherapy outcome prediction: A bladder cancer case study. In Pattern Recognition. ICPR International Workshops and Challenges: Virtual Event, January 10–15, 2021, Proceedings, Part I (pp. 227-242). Springer International Publishing.
Immunotherapy is one of the most interesting and promising cancer treatments. Encouraging results have confirmed the effectiveness of immunotherapy drugs for treating tumors in terms of long-term survival and a significant reduction in toxicity compared to more traditional chemotherapy approaches. However, the percentage of patients eligible for immunotherapy is rather small, and this is likely related to the limited knowledge of physiological mechanisms by which certain subjects respond to the treatment while others have no benefit. To address this issue, the authors propose an innovative approach based on the use of a non-linear cellular architecture with a deep downstream classifier for selecting and properly augmenting 2D features from chest-abdomen CT images toward improving outcome prediction. The proposed pipeline has been designed to make it usable over an innovative embedded Point of Care system. The authors report a case study of the proposed solution applied to a specific type of aggressive tumor, namely Metastatic Urothelial Carcinoma (mUC). The performance evaluation (overall accuracy close to 93%) confirms the proposed approach effectiveness.
3D Non-Local Neural Network: A Non-Invasive Biomarker for Immunotherapy Treatment Outcome Prediction. Case-Study: Metastatic Urothelial Carcinoma
 Rundo, F., Banna, G. L., Prezzavento, L., Trenta, F., Conoci, S., & Battiato, S. (2020). 3d non-local neural network: A non-invasive biomarker for immunotherapy treatment outcome prediction. case-study: Metastatic urothelial carcinoma. Journal of Imaging, 6(12), 133.
Rundo, F., Banna, G. L., Prezzavento, L., Trenta, F., Conoci, S., & Battiato, S. (2020). 3d non-local neural network: A non-invasive biomarker for immunotherapy treatment outcome prediction. case-study: Metastatic urothelial carcinoma. Journal of Imaging, 6(12), 133.
Immunotherapy is regarded as one of the most significant breakthroughs in cancer treatment. Unfortunately, only a small percentage of patients respond properly to the treatment. Moreover, to date, there are no efficient bio-markers able to early discriminate the patients eligible for this treatment. In order to help overcome these limitations, an innovative non-invasive deep pipeline, integrating Computed Tomography (CT) imaging, is investigated for the prediction of a response to immunotherapy treatment. We report preliminary results collected as part of a case study in which we validated the implemented method on a clinical dataset of patients affected by Metastatic Urothelial Carcinoma. The proposed pipeline aims to discriminate patients with high chances of response from those with disease progression. Specifically, the authors propose ad-hoc 3D Deep Networks integrating Self-Attention mechanisms in order to estimate the immunotherapy treatment response from CT-scan images and such hemato-chemical data of the patients. The performance evaluation (average accuracy close to 92%) confirms the effectiveness of the proposed approach as an immunotherapy treatment response biomarker.
Breast shape analysis
G. Catanuto, W. Taher, N. Rocco, F. Catalano, D. Allegra, F. Milotta, F. Stanco, G. Gallo, M.B. Nava. Breast Shape Analysis with Curvature Estimates and Principal Component Analysis for Cosmetic and Reconstructive Breast Surgery. Aesthetic Surgery Journal. 2018.
In this work we focus on digital shape analysis of breast models to assist breast surgeon for medical and surgical purposes. A clinical procedure for female breast digital scan is proposed. After a manual ROI definition through cropping, the meshes are automatically processed. The breasts are represented exploiting “bag of normal” representation, resulting in a 64-d descriptor. PCA is computed and the obtained first two principal components are used to plot the breasts shape into a 2D space. We show how the breasts subject to a surgery change their representation in this space and provide a cue about the error in this estimation. We believe that the proposed procedure represents a valid solution to evaluate the results of surgeries, since one of the most important goal of the specialists is to symmetrically reconstruct breasts and an objective tool to measure the result is currently missing.
G. Catanuto, A. Spano, A. Pennati, E. Riggio, G.M. Farinella, G. Impoco, S. Spoto, G. Gallo, M.B. Nava. Experimental methodology for digital breast shape analysis and objective surgical outcome evaluation. Journal of Plastic, Reconstructive & Aesthetic Surgery; 61; 3; 314-318. 2008.
Outcome evaluation in cosmetic and reconstructive surgery of the breast is commonly performed visually or employing bi-dimensional photography. The reconstructive process in the era of anatomical implants requires excellent survey capabilities that mainly rely on surgeon experience. In this paper we present a set of parameters to unambiguously estimate the shape of natural and reconstructed breast. A digital laser scanner was employed on seven female volunteers. A graphic depiction of curvature of the thoracic surface has been the most interesting result.
Objective analysis of simple kidney cysts from CT images
S. Battiato, G.M. Farinella, G. Gallo, O. Garretto, C. Privitera. Objective analysis of simple kidney cysts from CT images. Medical Measurements and Applications (MeMeA), IEEE International Workshop, 146-149. 2009.
Simple kidney cysts analysis from CT images is nowadays performed in a direct visual and hardly reproducible way. Computer-aided measurements of simple kidney cysts from CT images may help radiologists to accomplish an objective analysis of the clinical cases under observation. We proposed a semi-automatic segmentation algorithm for this task. Experiments performed on real datasets confirm the effectiveness and usefulness of the proposed method.
Neurofuzzy Segmentation of Microarray Images
S. Battiato, G.M. Farinella, G. Gallo, G. C. Guarnera. Neurofuzzy Segmentation of Microarray Images. Proceedings of IAPR International Conference on Pattern Recognition (ICPR), pp. 1-4. Tampa, Florida, USA, 2008.
We proposed a novel microarray segmentation strategy to separate background and foreground signals in microarray images making use of a neurofuzzy processing pipeline. In particular a Kohonen Self Organizing Map followed by a Fuzzy K-Mean classifier are employed to properly manage critical cases like saturated spot and spike noise. To speed up the overall process a Hilbert sampling is performed together with an ad-hoc analysis of statistical distribution of signals. Experiments confirm the validity of the proposed technique both in terms of measured and visual inspection quality.